3DView
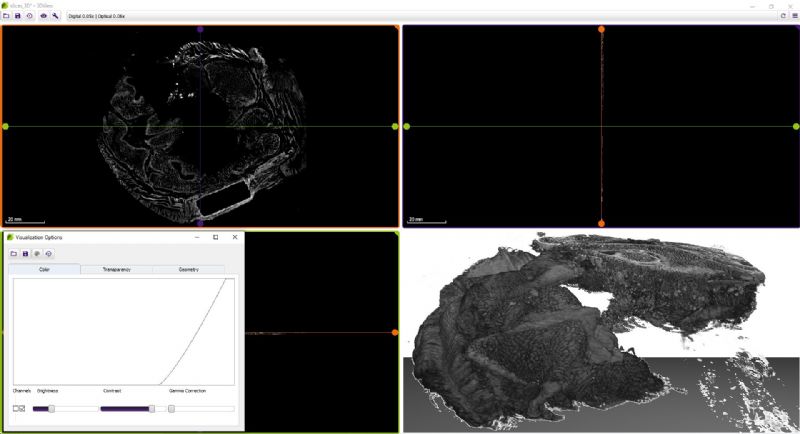
3DView by 3DHISTECH is a software application that prepares 3D reconstructions of 2D serial sections. It is also capable of displaying microCT images in 3D (VOL/VGI format).
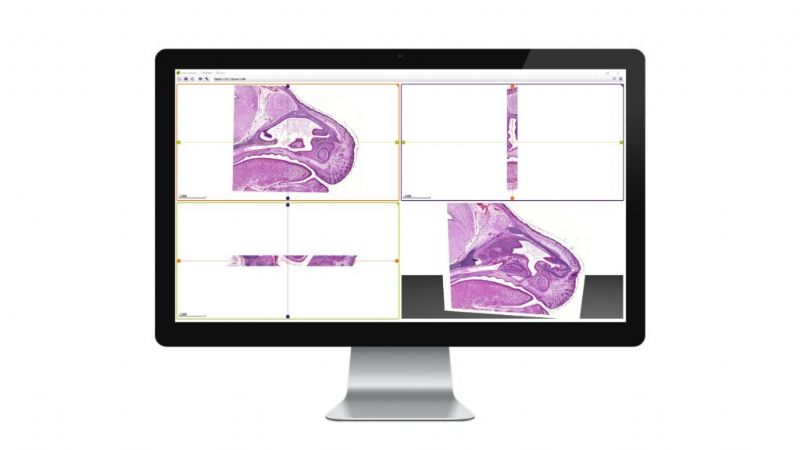
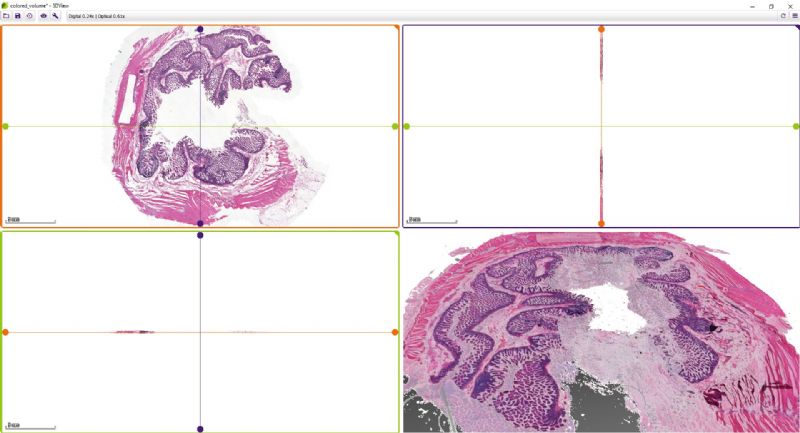
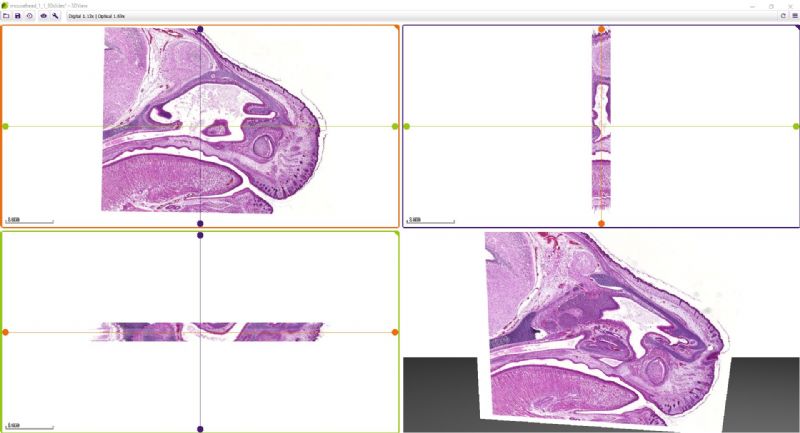
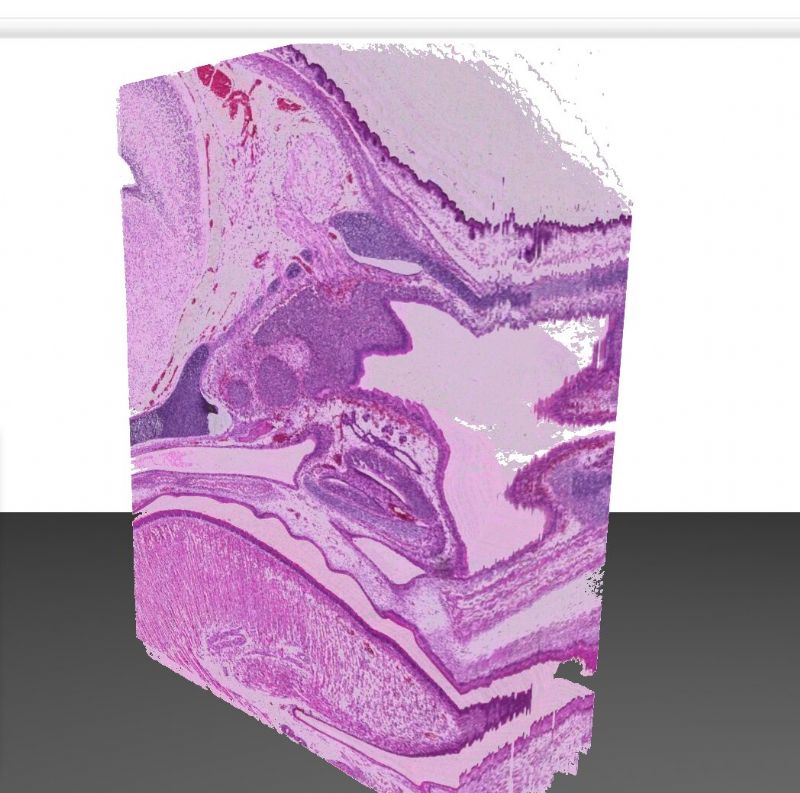
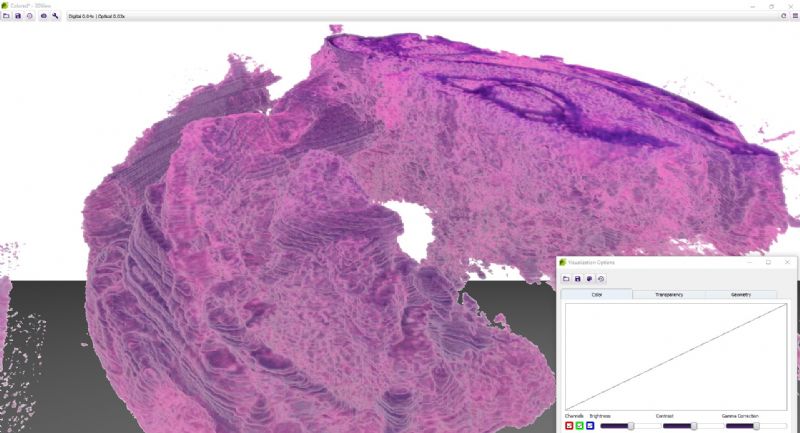
While microscope slides allow you to see only one section of reality, 3DView is a tool that can reconstruct the original tissue from its serial sections. The 3D image can then be rotated, zoomed into and “sliced” according to user needs.
How 3DView Works
- Aligns several 2D serial sections using the SlideMatch function;
- Prepares a 3D reconstruction from 2D serial sections;
- The 3D image can then be rotated, zoomed into and “sliced” according to user needs.
Key features
Advanced visualization options
- Besides brightness, contrast, gamma, RGB channels, etc., the user can select and edit several stainings for best visualization.
- Slice view (2D) of arbitrary sections and volume view (3D) of a volumetric data set are both available.
- 3DView is also capable of preparing 3D reconstructions from a Z-stack slide.
Measurement tools to assess metric distances
Advanced file export options
- The reconstruction can be exported as either 2D series or a 3D volume, with several channel data, alignment, export magnification and file format options – including high-resolution video.
- High-resolution snapshot export for publications is also available.
Wide compatibility with third-party file formats
Besides MRXS by 3DHISTECH, 3DView handles SVS by Leica Aperio, CZI by Zeiss, NDP by Hamamatsu, and VSI by Olympus.
Minimum system requirements
- Operating system: Windows 7 SP1, 8. 8.1 or 10 (64-bit version)
- Graphics processing unit: OpenGL 3.3 capable (with dedicated video memory of 512 MB minimum, 1 GB recommended), such as from AMD/ATI, Barco, Intel, or NVIDIA
- CPU: Intel Core i7 or comparable
- RAM: 4 GB minimum, 8 GB recommended